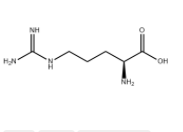
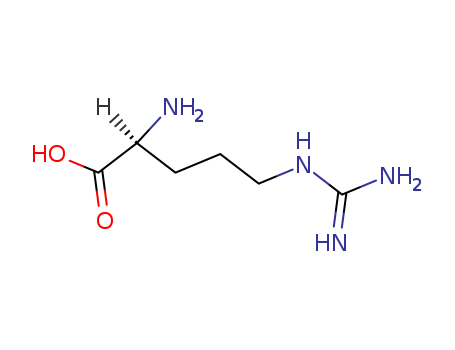
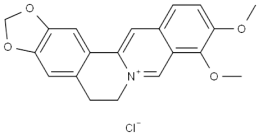
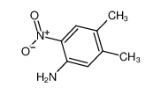
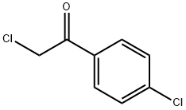
L(+)-Arginine literature
Direct monitoring of biocatalytic deacetylation of amino acid substrates by1H NMR reveals fine details of substrate specificity
De Cesare, Silvia,McKenna, Catherine A.,Mulholland, Nicholas,Murray, Lorna,Bella, Juraj,Campopiano, Dominic J.
supporting information, p. 4904 - 4909 (2021/06/16)
Amino acids are key synthetic building blocks that can be prepared in an enantiopure form by biocatalytic methods. We show that thel-selective ornithine deacetylase ArgE catalyses hydrolysis of a wide-range ofN-acyl-amino acid substrates. This activity was revealed by1H NMR spectroscopy that monitored the appearance of the well resolved signal of the acetate product. Furthermore, the assay was used to probe the subtle structural selectivity of the biocatalyst using a substrate that could adopt different rotameric conformations.
Argicyclamides A-C Unveil Enzymatic Basis for Guanidine Bis-prenylation
Balloo, Nandani,Fujita, Kei,Matsuda, Kenichi,Okino, Tatsufumi,Phan, Chin-Soon,Wakimoto, Toshiyuki
supporting information, p. 10083 - 10087 (2021/07/26)
Guanidine prenylation is an outstanding modification in alkaloid and peptide biosynthesis, but its enzymatic basis has remained elusive. We report the isolation of argicyclamides, a new class of cyanobactins with unique mono- and bis-prenylations on guanidine moieties, from Microcystis aeruginosa NIES-88. The genetic basis of argicyclamide biosynthesis was established by the heterologous expression and in vitro characterization of biosynthetic enzymes including AgcF, a new guanidine prenyltransferase. This study provides important insight into the biosynthesis of prenylated guanidines and offers a new toolkit for peptide modification.
Binding Methylarginines and Methyllysines as Free Amino Acids: A Comparative Study of Multiple Host Classes**
Bayer, Peter,Hof, Fraser,Isaacs, Lyle,Kamba, Bianca E.,Le, My-Hue,Schrader, Thomas,Warmerdam, Zoey
, (2021/11/30)
Methylated free amino acids are an important class of targets for host-guest chemistry that have recognition properties distinct from those of methylated peptides and proteins. We present comparative binding studies for three different host classes that are each studied with multiple methylated arginines and lysines to determine fundamental structure-function relationships. The hosts studied are all anionic and include three calixarenes, two acyclic cucurbiturils, and two other cleft-like hosts, a clip and a tweezer. We determined the binding association constants for a panel of methylated amino acids using indicator displacement assays. The acyclic cucurbiturils display stronger binding to the methylated amino acids, and some unique patterns of selectivity. The two other cleft-like hosts follow two different trends, shallow host (clip) following similar trends to the calixarenes, and the other more closed host (tweezer) binding certain less-methylated amino acids stronger than their methylated counterparts. Molecular modelling sheds some light on the different preferences of the various hosts. The results identify hosts with new selectivities and with affinities in a range that could be useful for biomedical applications. The overall selectivity patterns are explained by a common framework that considers the geometry, depth of binding pockets, and functional group participation across all host classes.
Sequence-Selective Protection of Peptides from Proteolysis
Li, Xiaowei,Chen, Kaiqian,Zhao, Yan
supporting information, p. 11092 - 11097 (2021/04/05)
Proteolysis of proteins and peptides is involved in the infection of cells by enveloped viruses and also in the invasion and spread of cancer cells. Shutting down broad-specificity proteases, however, is problematic because normal functions by these proteases will be affected. Herein, nanoparticle receptors were prepared from molecular imprinting for complex biological peptides. Their strong and selective binding enabled them to protect their targeted sequences from proteolysis in aqueous solution at stoichiometric amounts. Generality of the method was demonstrated by the protection of hydrophobic and hydrophilic peptides from different proteases, selective protection of a segment of a long peptide, and selective protection of a targeted peptide in a mixture. Most interestingly, two receptors targeting different parts of a long peptide could work in cooperation to protect the overall sequence, highlighting the versatility of the method.