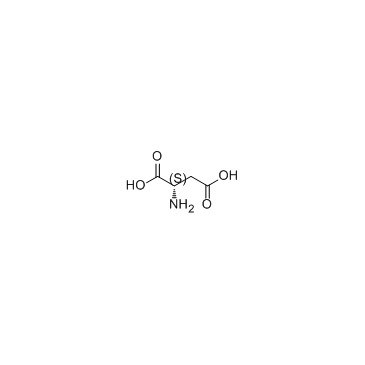
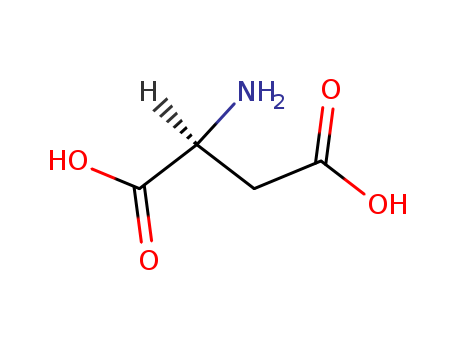
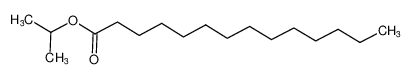
L-Aspartic acid literature
Structures and antitumor activities of ten new and twenty known surfactins from the deep-sea bacterium Limimaricola sp. SCSIO 53532
Chen, Min,Chen, Rouwen,Ding, Wenping,Li, Yanqun,Tian, Xinpeng,Yin, Hao,Zhang, Si
, (2022/01/11)
Surfactins are natural biosurfactants with myriad potential applications in the areas of healthcare and environment. However, surfactins were almost exclusively produced by the bacterium Bacillus species in previous reported literatures, together with difficulty in isolating pure monomer, which resulted in making extensive effort to remove duplication and little discovery of new surfactins in recent years. In the present study, the result of Molecular Networking indicated that Limimaricola sp. SCSIO 53532 might well be a potential resource for surfacin-like compounds based on OSMAC strategy. To search for new surfactins with significant biological activity, further study was undertaken on the strain. As a result, ten new surfactins (1–10), along with twenty known surfactins (11–30), were isolated from the ethyl acetate extract of SCSIO 53532. Their chemical structures were established by detailed 1D and 2D NMR spectroscopy, HRESIMS data, secondary ion mass spectrometry (MS/MS) analysis, and chemical degradation (Marfey's method) analysis. Cytotoxic activities of twenty-seven compounds against five human tumor cell lines were tested, and five compounds showed significant antitumor activities with IC50 values less than 10 μM. Furtherly, analysis of structure–activity relationships revealed that the branch of side chain, the esterification of Glu or Asp residue, and the amino acid residue of position 7 possessed a great influence on antitumor activity.
Recreating the natural evolutionary trend in key microdomains provides an effective strategy for engineering of a thermomicrobial N-demethylase
Gu, Zhenghua,Guo, Zitao,Shao, Jun,Shen, Chen,Shi, Yi,Tang, Mengwei,Xin, Yu,Zhang, Liang
, (2022/03/09)
N-demethylases have been reported to remove the methyl groups on primary or secondary amines, which could further affect the properties and functions of biomacromolecules or chemical compounds; however, the substrate scope and the robustness of N-demethylases have not been systematically investigated. Here we report the recreation of natural evolution in key microdomains of the Thermomicrobium roseum sarcosine oxidase (TrSOX), an N-demethylase with marked stability (melting temperature over 100 C) and enantioselectivity, for enhanced substrate scope and catalytic efficiency on -C-N-bonds. We obtained the structure of TrSOX by crystallization and X-ray diffraction (XRD) for the initial framework. The natural evolution in the nonconserved residues of key microdomains—including the catalytic loop, coenzyme pocket, substrate pocket, and entrance site—was then identified using ancestral sequence reconstruction (ASR), and the substitutions that accrued during natural evolution were recreated by site-directed mutagenesis. The single and double substitution variants catalyzed the N-demethylation of N-methyl-L-amino acids up to 1800- and 6000-fold faster than the wild type, respectively. Additionally, these single substitution variants catalyzed the terminal N-demethylation of non-amino-acid compounds and the oxidation of the main chain -C-N- bond to a -C=N- bond in the nitrogen-containing heterocycle. Notably, these variants retained the enantioselectivity and stability of the initial framework. We conclude that the variants of TrSOX are of great potential use in N-methyl enantiomer resolution, main-chain Schiff base synthesis, and alkaloid modification or degradation.
Leveraging Peptaibol Biosynthetic Promiscuity for Next-Generation Antiplasmodial Therapeutics
Lee, Jin Woo,Collins, Jennifer E.,Wendt, Karen L.,Chakrabarti, Debopam,Cichewicz, Robert H.
supporting information, p. 503 - 517 (2021/03/01)
Malaria remains a worldwide threat, afflicting over 200 million people each year. The emergence of drug resistance against existing therapeutics threatens to destabilize global efforts aimed at controlling Plasmodium spp. parasites, which is expected to leave vast portions of humanity unprotected against the disease. To address this need, systematic testing of a fungal natural product extract library assembled through the University of Oklahoma Citizen Science Soil Collection Program has generated an initial set of bioactive extracts that exhibit potent antiplasmodial activity (EC50 < 0.30 μg/mL) and low levels of toxicity against human cells (less than 50% reduction in HepG2 growth at 25 μg/mL). Analysis of the two top-performing extracts from Trichoderma sp. and Hypocrea sp. isolates revealed both contained chemically diverse assemblages of putative peptaibol-like compounds that were responsible for their antiplasmodial actions. Purification and structure determination efforts yielded 30 new peptaibols and lipopeptaibols (1-14 and 28-43), along with 22 known metabolites (15-27 and 44-52). While several compounds displayed promising activity profiles, one of the new metabolites, harzianin NPDG I (14), stood out from the others due to its noteworthy potency (EC50 = 0.10 μM against multi-drug-resistant P. falciparum line Dd2) and absence of gross toxicity toward HepG2 at the highest concentrations tested (HepG2 EC50 > 25 μM, selectivity index > 250). The unique chemodiversity afforded by these fungal isolates serves to unlock new opportunities for translating peptaibols into a bioactive scaffold worthy of further development.
Biosynthesis ofl-alanine fromcis-butenedioic anhydride catalyzed by a triple-enzyme cascadeviaa genetically modified strain
Cui, Ruizhi,Liu, Zhongmei,Yu, Puyi,Zhou, Li,Zhou, Zhemin
, p. 7290 - 7298 (2021/09/28)
In industry,l-alanine is biosynthesized using fermentation methods or catalyzed froml-aspartic acid by aspartate β-decarboxylase (ASD). In this study, a triple-enzyme system was developed to biosynthesizel-alanine fromcis-butenedioic anhydride, which was cost-efficient and could overcome the shortcomings of fermentation. Maleic acid formed bycis-butenedioic anhydride dissolving in water was transformed tol-alanineviafumaric acid andl-asparagic acid catalyzed by maleate isomerase (MaiA), aspartase (AspA) and ASD, respectively. The enzymatic properties of ASD from different origins were investigated and compared, as ASD was the key enzyme of the triple-enzyme cascade. Based on cofactor dependence and cooperation with the other two enzymes, a suitable ASD was chosen. Two of the three enzymes, MaiA and ASD, were recombinant enzymes cloned into a dual-promoter plasmid for overexpression; another enzyme, AspA, was the genomic enzyme of the host cell, in which AspA was enhanced by a T7promoter. Two fumarases in the host cell genome were deleted to improve the utilization of the intermediate fumaric acid. The conversion of whole-cell catalysis achieved 94.9% in 6 h, and the productivity given in our system was 28.2 g (L h)?1, which was higher than the productivity that had been reported. A catalysis-extraction circulation process for the synthesis ofl-alanine was established based on high-density fermentation, and the wastewater generated by this process was less than 34% of that by the fermentation process. Our results not only established a new green manufacturing process forl-alanine production fromcis-butenedioic anhydride but also provided a promising strategy that could consider both catalytic ability and cell growth burden for multi-enzyme cascade catalysis.